
An Introduction to t_TOST
A new function for TOST with t-tests
Aaron R. Caldwell
2026-01-26
IntroTOSTt.RmdIntroduction to TOST
TOST (Two One-Sided Tests) is a statistical approach used primarily for equivalence testing. Unlike traditional null hypothesis significance testing which aims to detect differences, equivalence testing aims to statistically support a claim of “no meaningful difference” between groups or conditions. This is particularly useful in bioequivalence studies, non-inferiority trials, or any research where establishing similarity (rather than difference) is the primary goal.
In an effort to make TOSTER more informative and easier
to use, I created the functions t_TOST and
simple_htest. These functions operate very similarly to
base R’s t.test function with a few key enhancements:
-
t_TOSTperforms 3 t-tests simultaneously:- One traditional two-tailed test
- Two one-sided tests to assess equivalence
-
simple_htestallows you to run equivalence testing or minimal effects testing using either t-tests or Wilcoxon-Mann-Whitney tests, with output in the familiarhtestclass format (like base R’s statistical test functions).
Both functions have versatile methods where you can supply:
- Two vectors of data
- A formula interface (e.g.,
y ~ group) - Various test configurations (two-sample, one-sample, or paired samples)
The functions also provide enhanced summary information and visualizations that make interpreting results more intuitive and user-friendly.
Beyond Equivalence: Minimal Effects Testing
These functions are not limited to equivalence tests. Minimal effects testing (MET) is also supported, which is useful for situations where the hypothesis concerns a minimal effect and the null hypothesis is equivalence. This reverses the typical TOST paradigm and can be valuable in contexts where you want to demonstrate that an effect exceeds some minimal threshold.
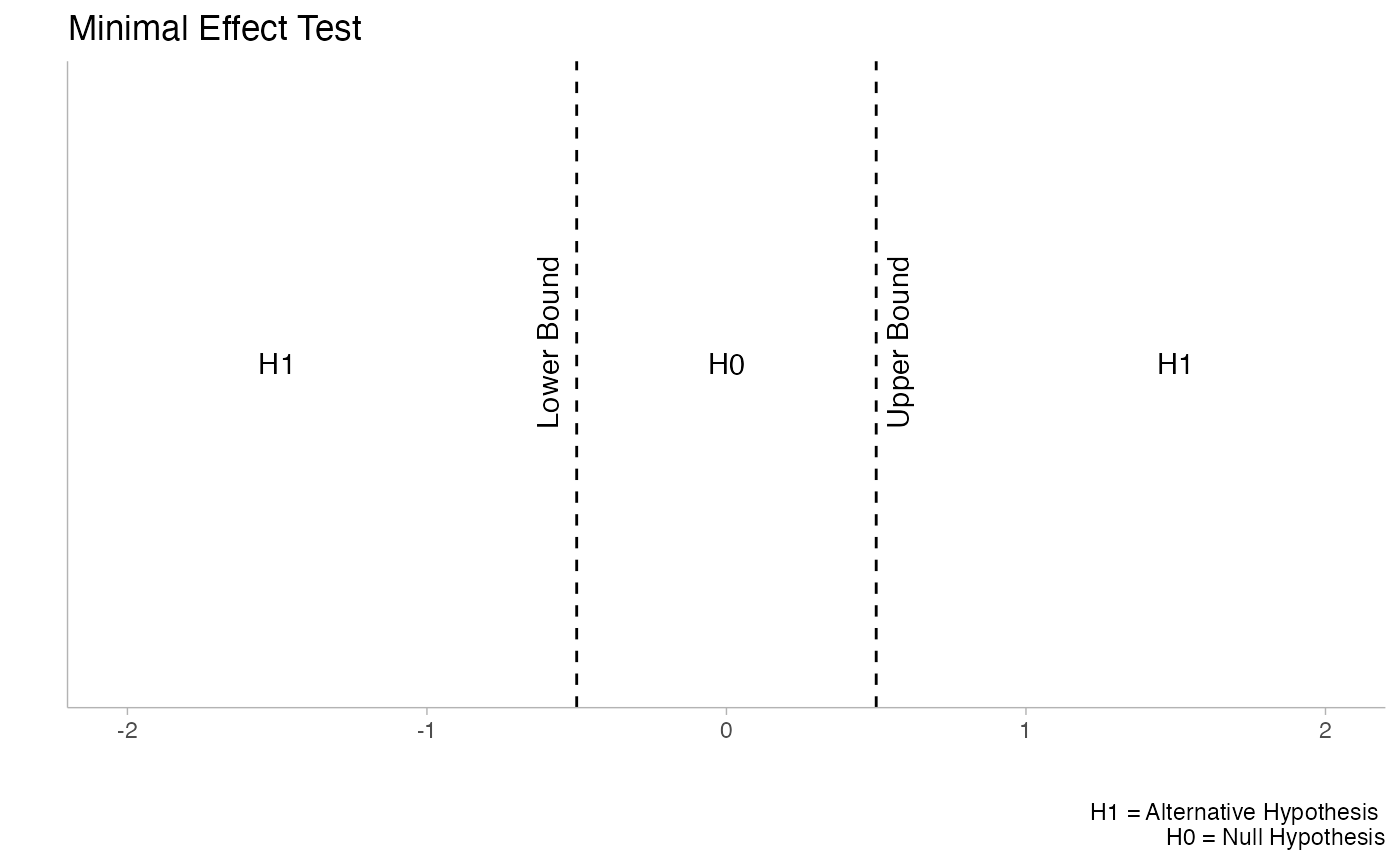
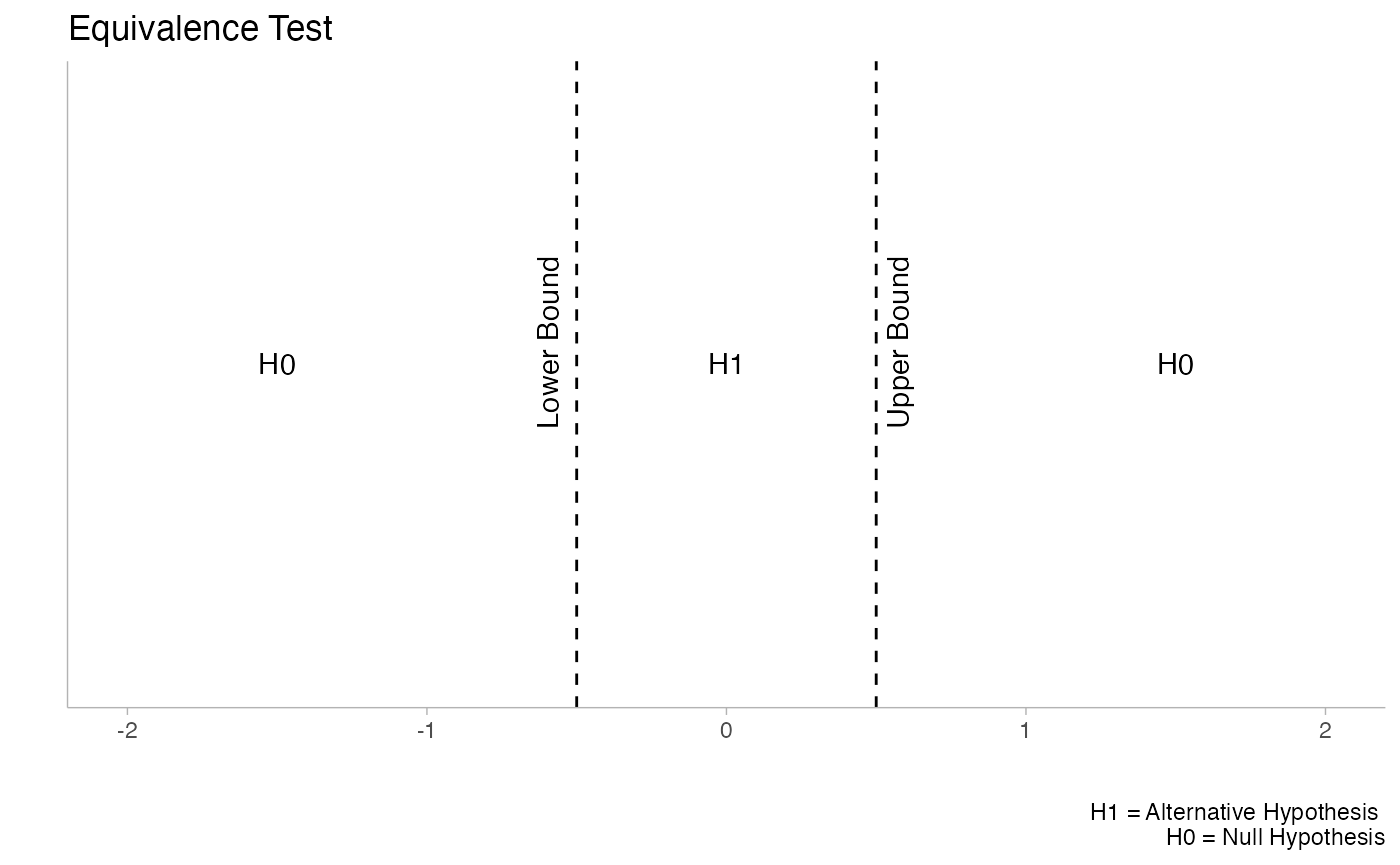
The plots above illustrate the conceptual difference between Minimal Effect Tests (left) and Equivalence Tests (right). Notice how the null (H0) and alternative (H1) hypotheses are reversed between these approaches.
Getting Started with Sample Data
In this vignette, we’ll use the built-in iris and
sleep datasets to demonstrate various applications of
TOST.
Let’s take a quick look at the sleep data:
head(sleep)
#> extra group ID
#> 1 0.7 1 1
#> 2 -1.6 1 2
#> 3 -0.2 1 3
#> 4 -1.2 1 4
#> 5 -0.1 1 5
#> 6 3.4 1 6The sleep dataset contains observations on the effect of
two soporific drugs (group 1 and 2) on 10 patients, with
the increase in hours of sleep (extra) as the response
variable.
Independent Groups Analysis
Basic Equivalence Testing
For our first example, we’ll test whether the effect of the two drugs
in the sleep data is equivalent within bounds of ±0.5
hours. We’ll use both the comprehensive t_TOST function and
the simpler simple_htest approach.
Using t_TOST
res1 = t_TOST(formula = extra ~ group,
data = sleep,
eqb = .5, # equivalence bounds of ±0.5 hours
smd_ci = "t") # t-distribution for SMD confidence intervals
# Alternative syntax with separate vectors
res1a = t_TOST(x = subset(sleep, group==1)$extra,
y = subset(sleep, group==2)$extra,
eqb = .5)The eqb parameter specifies our equivalence bounds (±0.5
hours of sleep). The smd_ci = "t" argument indicates that
we want to use the t-distribution and the standard error of the
standardized mean difference (SMD) to create SMD confidence intervals,
which is computationally faster for demonstration purposes.
Using simple_htest
We can achieve similar results with the more concise
simple_htest function:
# Simple htest approach
res1b = simple_htest(formula = extra ~ group,
data = sleep,
mu = .5, # equivalence bound
alternative = "e") # "e" for equivalenceThe alternative = "e" specifies an equivalence test, and
mu = .5 sets our equivalence bound.
Viewing Results
Let’s examine the output from both approaches:
# Comprehensive t_TOST output
print(res1)
#>
#> Welch Two Sample t-test
#>
#> The equivalence test was non-significant, t(17.78) = -1.3, p = 0.89
#> The null hypothesis test was non-significant, t(17.78) = -1.86, p = 0.08
#> NHST: don't reject null significance hypothesis that the effect is equal to zero
#> TOST: don't reject null equivalence hypothesis
#>
#> TOST Results
#> t df p.value
#> t-test -1.861 17.78 0.079
#> TOST Lower -1.272 17.78 0.890
#> TOST Upper -2.450 17.78 0.012
#>
#> Effect Sizes
#> Estimate SE C.I. Conf. Level
#> Raw -1.5800 0.8491 [-3.0534, -0.1066] 0.9
#> Hedges's g(av) -0.7965 0.4900 [-1.6467, 0.0537] 0.9
#> Note: SMD confidence intervals are an approximation. See vignette("SMD_calcs").
# Concise htest output
print(res1b)
#>
#> Welch Two Sample t-test
#>
#> data: extra by group
#> t = -1.2719, df = 17.776, p-value = 0.8901
#> alternative hypothesis: equivalence
#> null values:
#> difference in means difference in means
#> -0.5 0.5
#> 90 percent confidence interval:
#> -3.0533815 -0.1066185
#> sample estimates:
#> mean of x mean of y
#> 0.75 2.33Notice how t_TOST provides detailed information
including both raw and standardized effect sizes, whereas
simple_htest gives a more concise summary in the familiar
format of base R’s statistical tests.
Visualizing Results
One of the advantages of t_TOST is its built-in
visualization capabilities. There are four types of plots available:
1. Simple Dot-and-Whisker Plot
This is the default plot type, showing the point estimate and confidence intervals relative to the equivalence bounds:
plot(res1, type = "simple")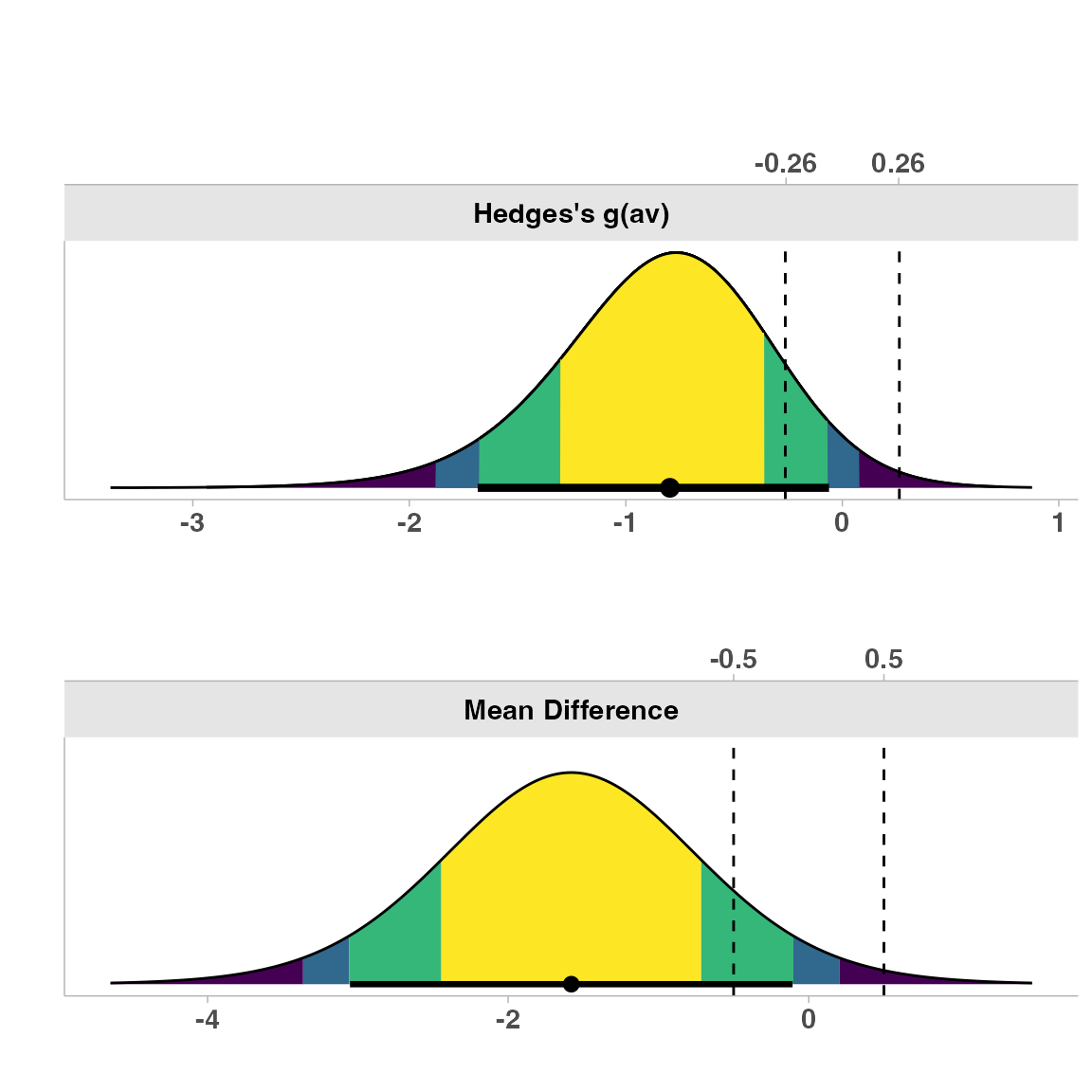
This plot clearly shows where our observed difference (with confidence intervals) falls in relation to our equivalence bounds (dashed vertical lines).
2. Consonance Density Plot
This plot shows the distribution of possible effect sizes, with shading for different confidence levels:
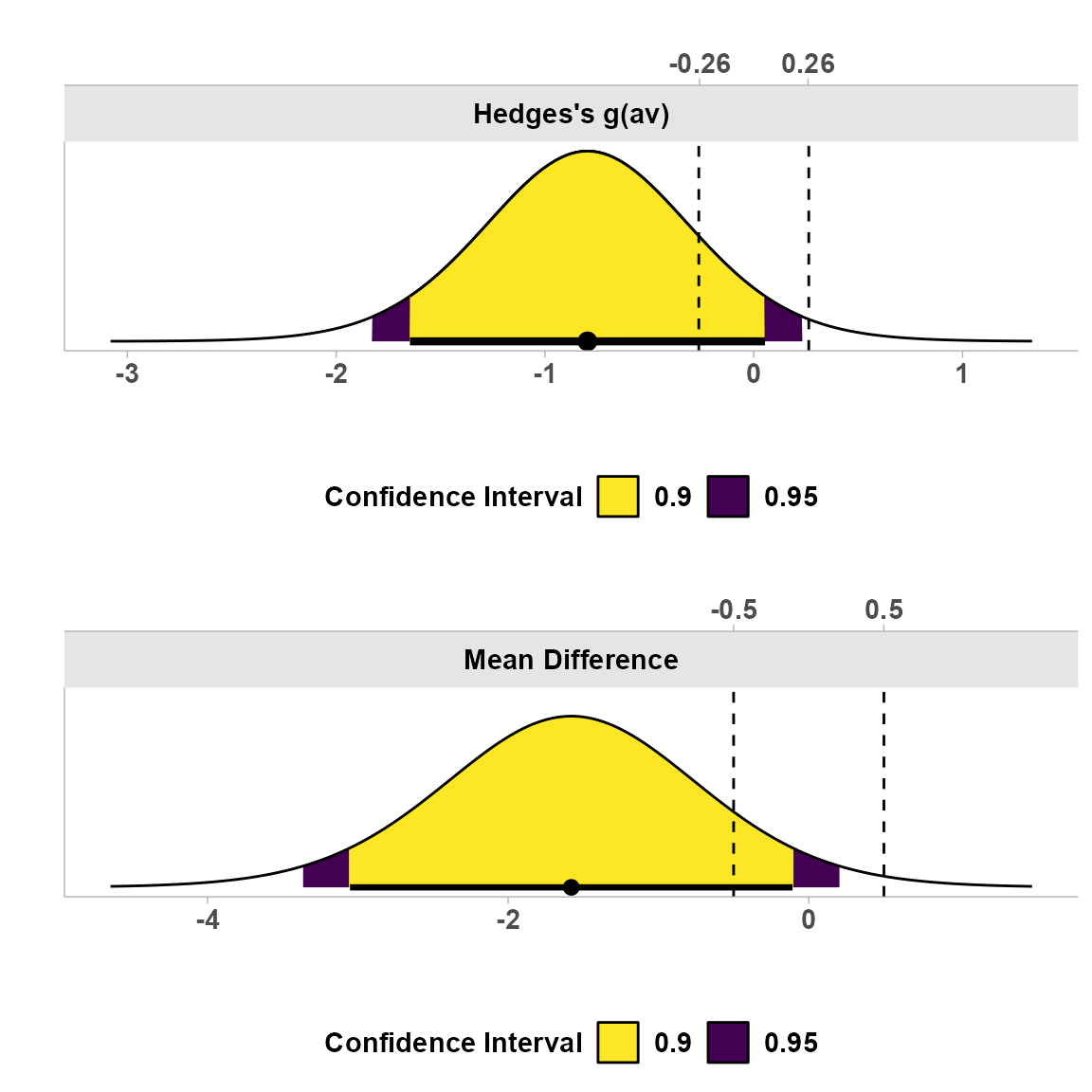
The darker shaded region represents the 90% confidence interval, while the lighter region represents the 95% confidence interval. This visualization helps illustrate the precision of our estimate.
3. Consonance Plot
This plot shows multiple confidence intervals simultaneously:
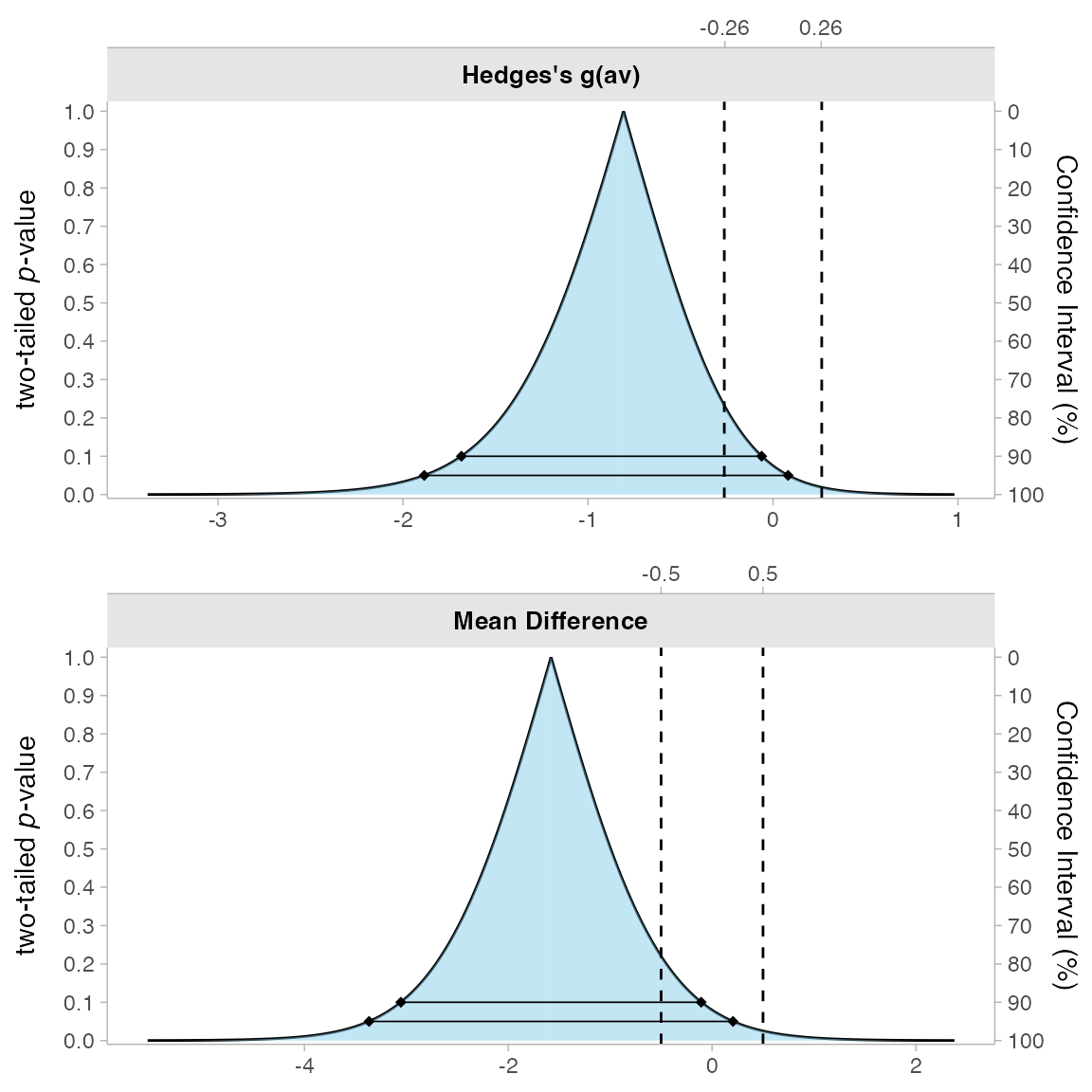
Here, we can see both the 90% and 95% confidence intervals represented as different lines.
4. Null Distribution Plot
This plot visualizes the null distribution:
plot(res1, type = "tnull")
#> SMD cannot be plotted if type = "tnull"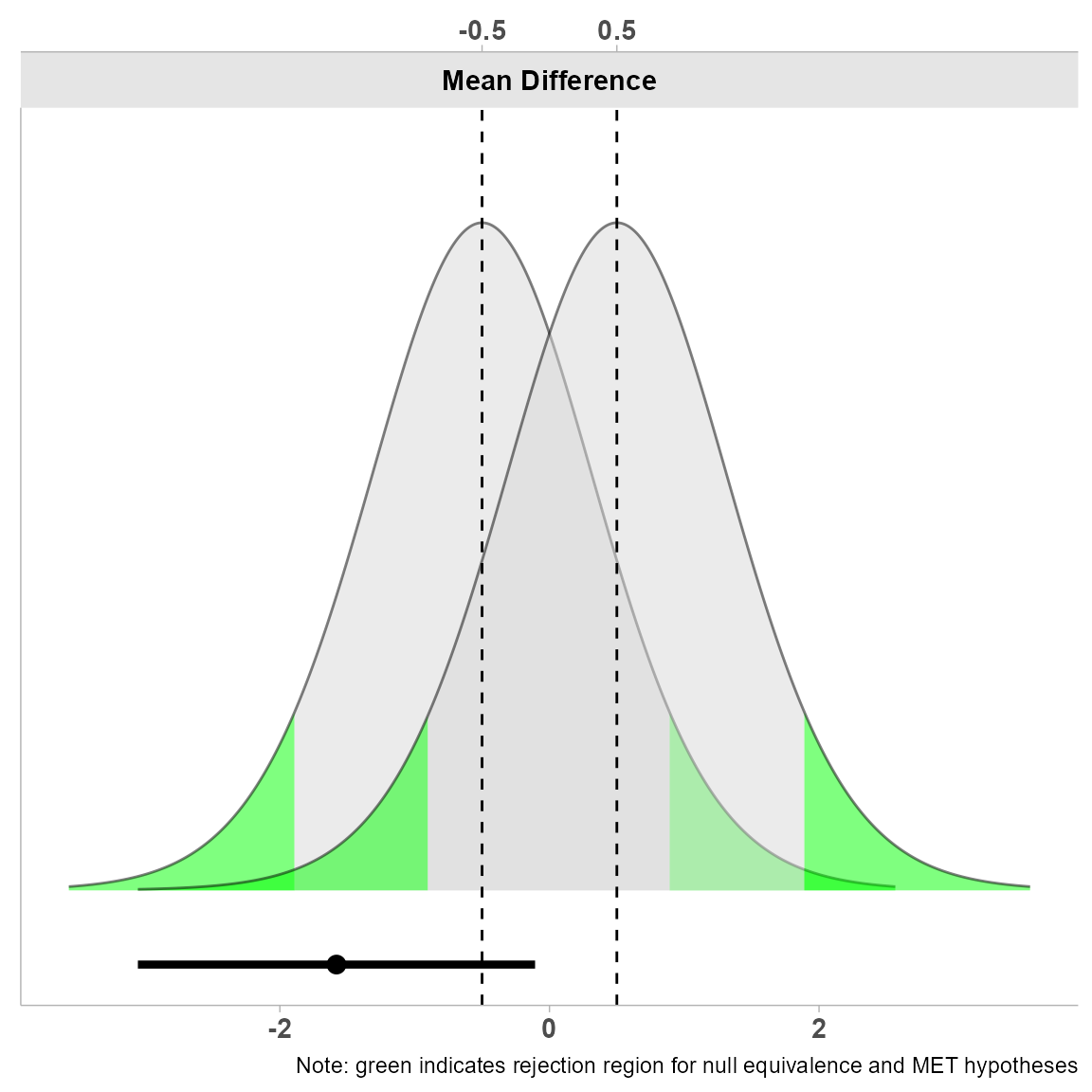
This visualization is particularly useful for understanding the theoretical distribution under the null hypothesis.
Describing Results in Plain Language
Both t_TOST and simple_htest objects can be
used with the describe or describe_htest
functions to generate plain language interpretations of the results:
describe(res1)
describe_htest(res1b)Using the Welch Two Sample t-test, a null hypothesis significance test (NHST), and a equivalence test, via two one-sided tests (TOST), were performed with an alpha-level of 0.05. These tested the null hypotheses that true mean difference is equal to 0 (NHST), and true mean difference is more extreme than -0.5 and 0.5 (TOST). Both the equivalence test (p = 0.89), and the NHST (p = 0.079) were not significant (mean difference = -1.58 90% C.I.[-3.05, -0.107]; Hedges’s g(av) = -0.796 90% C.I.[-1.65, 0.054]). Therefore, the results are inconclusive: neither null hypothesis can be rejected.
The Welch Two Sample t-test is not statistically significant (t(17.776) = -1.27, p = 0.89, mean of x = 0.75, mean of y = 2.33, 90% C.I.[-3.05, -0.107]) at a 0.05 alpha-level. The null hypothesis cannot be rejected. At the desired error rate, it cannot be stated that the true difference in means is between -0.5 and 0.5.
These descriptions provide accessible interpretations of the statistical results, making it easier to understand and communicate findings.
Paired Samples Analysis
Many study designs involve paired measurements (e.g., before-after designs or matched samples). Let’s examine how to perform TOST with paired data.
Using the Sleep Data with Paired Analysis
We can use the same sleep data, but now treat the measurements as paired:
# For paired tests, use separate vectors rather than formula notation
res2 = t_TOST(x = sleep$extra[sleep$group == 1],
y = sleep$extra[sleep$group == 2],
paired = TRUE, # specify paired analysis
eqb = .5)
res2
#>
#> Paired t-test
#>
#> The equivalence test was non-significant, t(9) = -2.8, p = 0.99
#> The null hypothesis test was significant, t(9) = -4.06, p < 0.01
#> NHST: reject null significance hypothesis that the effect is equal to zero
#> TOST: don't reject null equivalence hypothesis
#>
#> TOST Results
#> t df p.value
#> t-test -4.062 9 0.003
#> TOST Lower -2.777 9 0.989
#> TOST Upper -5.348 9 < 0.001
#>
#> Effect Sizes
#> Estimate SE C.I. Conf. Level
#> Raw -1.580 0.3890 [-2.293, -0.867] 0.9
#> Hedges's g(z) -1.174 0.4412 [-1.8046, -0.4977] 0.9
#> Note: SMD confidence intervals are an approximation. See vignette("SMD_calcs").
res2b = simple_htest(
x = sleep$extra[sleep$group == 1],
y = sleep$extra[sleep$group == 2],
paired = TRUE,
mu = .5,
alternative = "e")
res2b
#>
#> Paired t-test
#>
#> data: x and y
#> t = -2.7766, df = 9, p-value = 0.9892
#> alternative hypothesis: equivalence
#> null values:
#> mean difference mean difference
#> -0.5 0.5
#> 90 percent confidence interval:
#> -2.2930053 -0.8669947
#> sample estimates:
#> mean difference
#> -1.58Setting paired = TRUE changes the analysis to account
for within-subject correlations. Note that for paired tests, we use
separate vectors (x and y) rather than the formula notation, as formula
notation represents independent groups.
Using Separate Vectors for Paired Data
Alternatively, we can provide two separate vectors for paired data, as demonstrated with the iris dataset:
res3 = t_TOST(x = iris$Sepal.Length,
y = iris$Sepal.Width,
paired = TRUE,
eqb = 1)
res3
#>
#> Paired t-test
#>
#> The equivalence test was non-significant, t(149) = 22.32, p = 1
#> The null hypothesis test was significant, t(149) = 34.815, p < 0.01
#> NHST: reject null significance hypothesis that the effect is equal to zero
#> TOST: don't reject null equivalence hypothesis
#>
#> TOST Results
#> t df p.value
#> t-test 34.82 149 < 0.001
#> TOST Lower 47.31 149 < 0.001
#> TOST Upper 22.32 149 1
#>
#> Effect Sizes
#> Estimate SE C.I. Conf. Level
#> Raw 2.786 0.08002 [2.6536, 2.9184] 0.9
#> Hedges's g(z) 2.828 0.18393 [2.5252, 3.1244] 0.9
#> Note: SMD confidence intervals are an approximation. See vignette("SMD_calcs").
res3a = simple_htest(
x = iris$Sepal.Length,
y = iris$Sepal.Width,
paired = TRUE,
mu = 1,
alternative = "e"
)
res3a
#>
#> Paired t-test
#>
#> data: x and y
#> t = 22.319, df = 149, p-value = 1
#> alternative hypothesis: equivalence
#> null values:
#> mean difference mean difference
#> -1 1
#> 90 percent confidence interval:
#> 2.653551 2.918449
#> sample estimates:
#> mean difference
#> 2.786Here we’re testing whether the difference between Sepal.Length and Sepal.Width is equivalent within ±1 unit.
Minimal Effect Testing (MET)
As mentioned earlier, sometimes we want to test for a minimal effect
rather than equivalence. We can do this by changing the
hypothesis argument to “MET”:
res_met = t_TOST(x = iris$Sepal.Length,
y = iris$Sepal.Width,
paired = TRUE,
hypothesis = "MET", # Change to minimal effect test
eqb = 1,
smd_ci = "t")
res_met
#>
#> Paired t-test
#>
#> The minimal effect test was significant, t(149) = 47.31, p < 0.01
#> The null hypothesis test was significant, t(149) = 34.815, p < 0.01
#> NHST: reject null significance hypothesis that the effect is equal to zero
#> TOST: reject null MET hypothesis
#>
#> TOST Results
#> t df p.value
#> t-test 34.82 149 < 0.001
#> TOST Lower 47.31 149 1
#> TOST Upper 22.32 149 < 0.001
#>
#> Effect Sizes
#> Estimate SE C.I. Conf. Level
#> Raw 2.786 0.08002 [2.6536, 2.9184] 0.9
#> Hedges's g(z) 2.828 0.30870 [2.3174, 3.3393] 0.9
#> Note: SMD confidence intervals are an approximation. See vignette("SMD_calcs").
res_metb = simple_htest(x = iris$Sepal.Length,
y = iris$Sepal.Width,
paired = TRUE,
mu = 1,
alternative = "minimal.effect")
res_metb
#>
#> Paired t-test
#>
#> data: x and y
#> t = 22.319, df = 149, p-value < 2.2e-16
#> alternative hypothesis: minimal.effect
#> null values:
#> mean difference mean difference
#> -1 1
#> 90 percent confidence interval:
#> 2.653551 2.918449
#> sample estimates:
#> mean difference
#> 2.786For simple_htest, we use
alternative = "minimal.effect" to specify MET. The
smd_ci = "t" in t_TOST specifies using the
t-distribution method for calculating standardized mean difference
confidence intervals (this is faster than the default “nct” method).
Interpreting MET Results
describe(res_met)
describe_htest(res_metb)Using the Paired t-test, a null hypothesis significance test (NHST), and a minimal effect test, via two one-sided tests (TOST), were performed with an alpha-level of 0.05. These tested the null hypotheses that true mean difference is equal to 0 (NHST), and true mean difference is greater than -1 or less than 1 (TOST). The minimal effect test was significant, t(149) = 22.319, p < 0.001 (mean difference = 2.786 90% C.I.[2.654, 2.918]; Hedges’s g(z) = 2.828 90% C.I.[2.317, 3.339]). At the desired error rate, it can be stated that the true mean difference is less than -1 or greater than 1.
The Paired t-test is statistically significant (t(149) = 22.319, p < 0.001, mean difference = 2.786, 90% C.I.[2.654, 2.918]) at a 0.05 alpha-level. The null hypothesis can be rejected. At the desired error rate, it can be stated that the true mean difference is less than -1 or greater than 1.
Warning: careful using narrow equivalence bounds
Now, using MET isn’t without some potential danger. If a user utilizes a very small equivalence bound (e.g., an SMD of 0.05) then there is the possibility that the p-value for the MET test will be smaller than the a typical nil-hypothesis two-tailed test. This raises the concern that someone could “hack” these functions in order to produce a “significant” result that alluded them when using a two-tailed test. A warning message has now been added to the TOST functions in this package to at least warn users about the hazards of using very narrow equivalence bounds.
set.seed(221)
dat1 = rnorm(30)
dat2 = rnorm(30)
test = t_TOST(
x = dat1,
y = dat2,
eqbound_type = "SMD",
eqb = .2,
hypothesis = "MET"
)
#> Warning: setting bound type to SMD produces biased results!
#> MET test may have higher error rates than a nil two-tailed test. Consider wider equivalence bounds.
test
#>
#> Welch Two Sample t-test
#>
#> The minimal effect test was non-significant, t(54.91) = 0.91, p = 0.74
#> The null hypothesis test was non-significant, t(54.91) = 0.133, p = 0.89
#> NHST: don't reject null significance hypothesis that the effect is equal to zero
#> TOST: don't reject null MET hypothesis
#>
#> TOST Results
#> t df p.value
#> t-test 0.1329 54.91 0.895
#> TOST Lower 0.9075 54.91 0.816
#> TOST Upper -0.6417 54.91 0.738
#>
#> Effect Sizes
#> Estimate SE C.I. Conf. Level
#> Raw 0.03003 0.2259 [-0.348, 0.408] 0.9
#> Hedges's g(av) 0.03385 0.2626 [-0.3852, 0.4526] 0.9
#> Note: SMD confidence intervals are an approximation. See vignette("SMD_calcs").One Sample t-test
In some cases, we may want to compare a single sample against known bounds. For this, we can use a one-sample test:
res4 = t_TOST(x = iris$Sepal.Length,
hypothesis = "EQU",
eqb = c(5.5, 8.5), # lower and upper bounds
smd_ci = "t")
#> Equivalence interval does not include zero.
res4
#>
#> One Sample t-test
#>
#> The equivalence test was significant, t(149) = 5.08, p < 0.01
#> The null hypothesis test was significant, t(149) = 86.425, p < 0.01
#> NHST: reject null significance hypothesis that the effect is equal to zero
#> TOST: reject null equivalence hypothesis
#>
#> TOST Results
#> t df p.value
#> t-test 86.425 149 < 0.001
#> TOST Lower 5.078 149 < 0.001
#> TOST Upper -39.293 149 < 0.001
#>
#> Effect Sizes
#> Estimate SE C.I. Conf. Level
#> Raw 5.843 0.06761 [5.7314, 5.9552] 0.9
#> Hedges's g 7.021 0.41350 [6.2039, 7.8381] 0.9
#> Note: SMD confidence intervals are an approximation. See vignette("SMD_calcs").Here, we’re testing whether the mean of Sepal.Length is equivalent within bounds of 5.5 and 8.5 units. Notice how we specify the bounds as a vector with two distinct values rather than a single value that gets applied symmetrically.
Working with Summary Statistics Only
Researchers often only have access to summary statistics rather than
raw data, especially when working with published literature. The
tsum_TOST function allows you to perform TOST analyses
using just summary statistics:
res_tsum = tsum_TOST(
m1 = mean(iris$Sepal.Length, na.rm=TRUE), # sample mean
sd1 = sd(iris$Sepal.Length, na.rm=TRUE), # sample standard deviation
n1 = length(na.omit(iris$Sepal.Length)), # sample size
hypothesis = "EQU",
eqb = c(5.5, 8.5)
)
#> Equivalence interval does not include zero.
res_tsum
#>
#> One-sample t-test
#>
#> The equivalence test was significant, t(149) = 5.078, p = 5.62e-07
#> The null hypothesis test was significant, t(149) = 86.425, p = 3.33e-129
#> NHST: reject null significance hypothesis that the effect is equal to zero
#> TOST: reject null equivalence hypothesis
#>
#> TOST Results
#> t df p.value
#> t-test 86.425 149 < 0.001
#> TOST Lower 5.078 149 < 0.001
#> TOST Upper -39.293 149 < 0.001
#>
#> Effect Sizes
#> Estimate SE C.I. Conf. Level
#> Raw 5.843 0.06761 [5.7314, 5.9552] 0.9
#> Hedges's g 7.021 0.41350 [6.327, 7.6914] 0.9
#> Note: SMD confidence intervals are an approximation. See vignette("SMD_calcs").Required arguments vary depending on the test type: *
n1 & n2: the sample sizes (only n1 needed for
one-sample tests) * m1 & m2: the sample means *
sd1 & sd2: the sample standard deviations *
r12: the correlation between paired samples (only needed if
paired = TRUE)
The same visualization and description methods work with
tsum_TOST:
plot(res_tsum)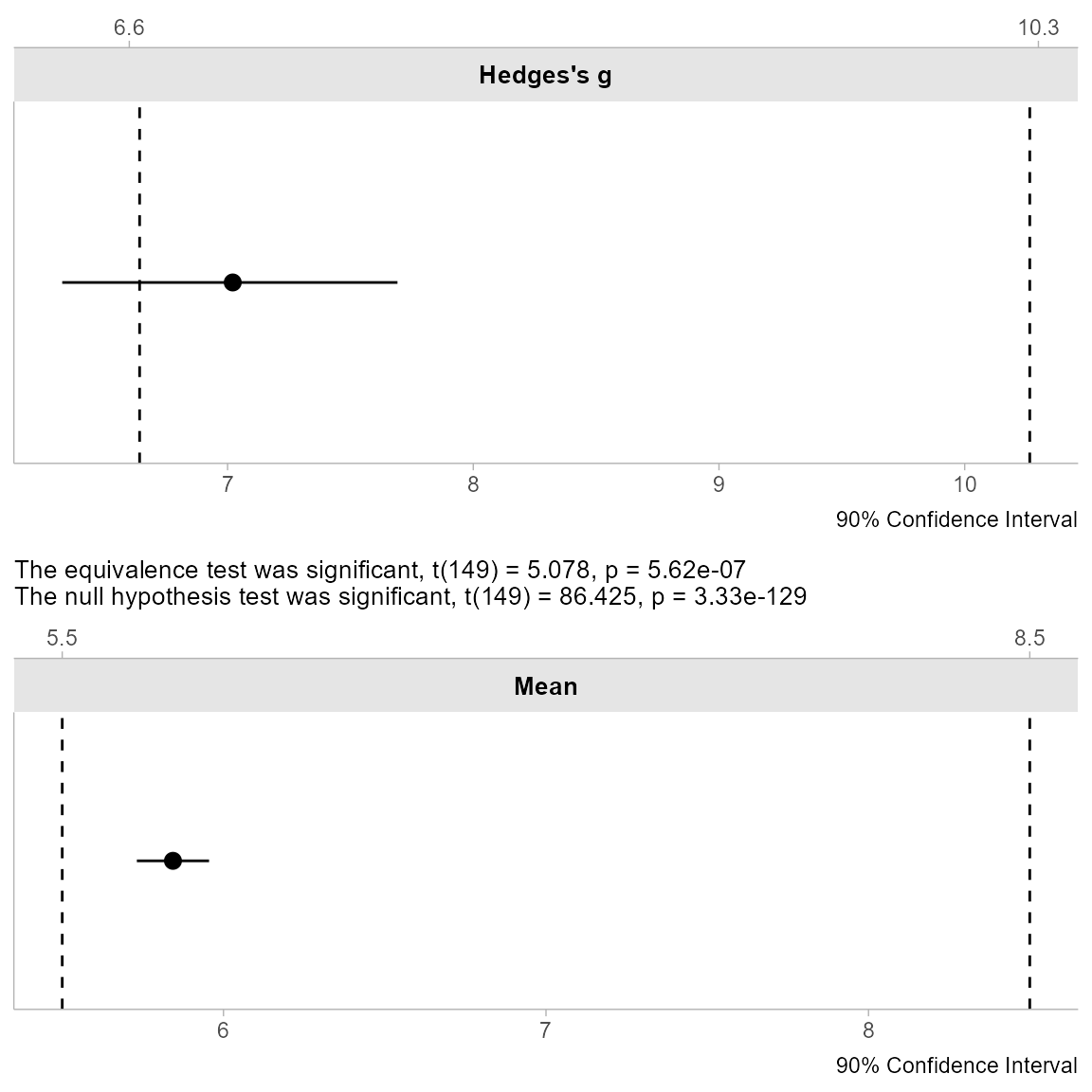
describe(res_tsum)
#> [1] "Using the One-sample t-test, a null hypothesis significance test (NHST), and a equivalence test, via two one-sided tests (TOST), were performed with an alpha-level of 0.05. These tested the null hypotheses that true mean is equal to 0 (NHST), and true mean is more extreme than 5.5 and 8.5 (TOST). The equivalence test was significant, t(149) = 5.078, p < 0.001 (mean = 5.843 90% C.I.[5.731, 5.955]; Hedges's g = 7.021 90% C.I.[6.327, 7.691]). At the desired error rate, it can be stated that the true mean is between 5.5 and 8.5."Power Analysis for TOST
Planning studies requires determining appropriate sample sizes. The
power_t_TOST function allows for power calculations
specifically designed for TOST analyses.
This function uses more accurate methods than previous TOSTER functions and matches results from commercial software like PASS. The calculations are based on Owen’s Q-function or direct integration of the bivariate non-central t-distribution1. Approximate power is implemented via the non-central t-distribution or the ‘shifted’ central t-distribution Diletti, Hauschke, and Steinijans (1992).
The interface mimics base R’s power.t.test function. You
specify equivalence bounds and leave one parameter blank
(alpha, power, or n) to solve for
it:
power_t_TOST(n = NULL,
delta = 1, # assumed true difference
sd = 2.5, # assumed standard deviation
eqb = 2.5, # equivalence bounds
alpha = .025, # significance level
power = .95, # desired power
type = "two.sample") # test type
#>
#> Two-sample TOST power calculation
#>
#> power = 0.95
#> beta = 0.05
#> alpha = 0.025
#> n = 73.16747
#> delta = 1
#> sd = 2.5
#> bounds = -2.5, 2.5
#>
#> NOTE: n is number in *each* groupIn this example, we’re planning a two-sample equivalence study where: - We assume the true difference is at least 1 unit - The standard deviation is estimated to be 2.5 - We set equivalence bounds to ±2.5 units - We want 95% power with alpha of 0.025
The analysis indicates we need 74 participants per group (148 total) to achieve our desired power.
For more advanced power analysis options, the PowerTOST
R package provides additional functionality.
Conclusion
The t_TOST and simple_htest functions in
the TOSTER package provide flexible and user-friendly tools for
performing equivalence testing and minimal effect testing. With support
for various study designs, robust visualization capabilities, and plain
language interpretations, these functions make it easier to apply and
communicate these important statistical approaches.
Whether you’re analyzing raw data or working with summary statistics, TOSTER provides the tools needed to conduct and interpret these tests appropriately.