exp(log(95)-log(100))[1] 0.95# to get inverse
1/exp(log(95)-log(100))[1] 1.052632Last Update: 2024-06-04
The physiology literature’s overuse of percentage change is problematic. While well intentioned, this approach is statistically flawed (see T. J. Cole 2000; Curran-Everett and Williams 2015; T. J. Cole and Altman 2017; Kaiser 1989). A key issue is the false symmetry it creates: a 10% weight loss followed by a 10% gain doesn’t equal the original weight.
Researchers likely use percentage change for its intuitive appeal and ease of comparison across studies. Most people would state they intuitively understand that a 5% increase in something is modest and a 25% increase in something is rather large. Additionally, there is the added benefit that different outcomes can be compared on a more universal scale (much like a standardized mean difference, e.g., Cohen’s d), and evidence for treatments/interventions could be synthesized in something like a meta-analysis (Friedrich, Adhikari, and Beyene 2011; Hedges, Gurevitch, and Curtis 1999).
Alternatives exist that avoid the pitfalls of typical percentage change/difference. Let’s explore these options.
I want to present the “sympercent” as T. J. Cole (2000) calls it. In his original paper, T. J. Cole (2000) presents the simple transformation of continuous variables (\(x\)) using the natural logarithm:
\[ log_ex \]
The symmetric percent difference/change (\(s\%\)) can then be calculated by multiplying the log transformed differences/changes by 100.
\[ s\% = 100 \cdot (log(x) -log(y)) \]
The natural log transformation offers variance stabilization, addressing heteroscedasticity concerns. Additionally, it provides intuitive interpretation. For example, in height differences between British adult males (177.3 cm) and females (163.6 cm), the log transformation yields a symmetric difference of 8.04%, aligning with our perception of a modest height difference (T. J. Cole 2000, e.g., \(100 \cdot (log(177.3) - log(163.6)) = 8.04 \space s\%\)).
Pre-post intervention studies often assess changes in biological variables. Instead of percentage change (\(\%change = \frac{post-pre}{pre}\)), sympercents (\(s\% = 100 \cdot (log(post) -log(pre))\)) offer a symmetric alternative with similar interpretation. For instance, a weight loss from 100 kg to 95 kg yields a 5.1 s% sympercent change, comparable to the 5% change, but without the asymmetry issue.
What is often done, and what I often do, is transform the sympercent to a ratio (I’ve also seen this referred to as a “factor” difference). This can be accomplished because the following is true.
\[ log(x) - log(y) = log(\frac{x}{y}) \]
Therefore, the ratio (\(\frac{x}{y}\)) can be obtained by taken by the exponentiation of the log transformed difference.
\[ ratio = e^{log(x)-log(y)} \] To use our example of weight loss earlier, we can the ratio the following way in R.
exp(log(95)-log(100))[1] 0.95# to get inverse
1/exp(log(95)-log(100))[1] 1.052632We could then interpret the data as showing a reduction in body weight by a factor of 1.05.
Sometimes I’ve seen people use the log transformation but then use the ratio to calculate a percentage. Essentially the producedure is as follows:
\[ ratio\% = 100*e^{log(x)-log(y)}-100 \]
Again, to use the example from before we get about a 5 percent decrease… However, as you can see from the results below the difference is no longer symmetric.
100*exp(log(95)-log(100))-100[1] -5# to get inverse
100*1/exp(log(95)-log(100))-100[1] 5.263158I think this is less than ideal, but so long as you are explicit in your methods this is fine as descriptive statistic in my view (i.e., it makes the results of log-transformed data/model more interpretable).
Let’s use a quick example to show how well the percent change, sympercent, and ratio percent track with each other.
Let’s use the weight loss study example from before and create a scenario where the average weight at baseline is 100kg with an SD of 10. I created a random sample below and calculated the different percent for each pair of observations
library(tidyverse)
x = rnorm(10,100,10)
y = rnorm(10,100,10)
df = data.frame(
x,
y
)
# Percent Change
df$pc = (x-y)/x
# Sympercent
df$sym = log(x)-log(y)
# Ratio Percent
df$rp = exp(df$sym)-1
df %>% flextable::flextable() %>% flextable::theme_tron()x | y | pc | sym | rp |
|---|---|---|---|---|
91.97173 | 114.65641 | -0.24664841 | -0.22045868 | -0.19784922 |
127.03798 | 105.04233 | 0.17314231 | 0.19012268 | 0.20939796 |
97.56966 | 105.16089 | -0.07780316 | -0.07492486 | -0.07218680 |
92.70250 | 102.91963 | -0.11021418 | -0.10455295 | -0.09927290 |
108.83737 | 98.95604 | 0.09078986 | 0.09517903 | 0.09985575 |
111.32794 | 113.03202 | -0.01530676 | -0.01519080 | -0.01507600 |
103.40234 | 98.53781 | 0.04704471 | 0.04818730 | 0.04936718 |
93.22710 | 95.15184 | -0.02064567 | -0.02043544 | -0.02022805 |
96.85504 | 91.23349 | 0.05804088 | 0.05979340 | 0.06161719 |
109.55669 | 97.62742 | 0.10888676 | 0.11528377 | 0.12219184 |
As we can see in this example, the sympercent closely approximates the percentage change; so does the ratio percent.
The sympercent suggestion by T. J. Cole (2000) is effective and easy to use. It avoids the downsides of traditional percentage difference/change, offering easier interpretation. Notably, the standard error remains on the same scale when reporting sympercent (unlike ratios). While model assumptions should always be checked, many biological/physiological variables are well-suited for this method
Below, I’m going to demonstrate some examples of using this type of analysis. One for a between-subjects design, another for repeated measures design, and lastly for a two-arm pre-post test design.
Let’s look at the Tobacco data from R package “PairedData”. To quote the documentation:
This dataset presents 8 paired data corresponding to numbers of lesions caused by two virus preparations inoculated into the two halves of each tobacco leaves.
library(tidyverse)
library(PairedData)
data(Tobacco)
tb = Tobacco %>%
janitor::clean_names() %>%
as_tibble()
tb# A tibble: 8 × 3
plant preparation_1 preparation_2
<fct> <int> <int>
1 P5 31 18
2 P6 20 17
3 P7 18 14
4 P3 17 11
5 P1 9 10
6 P4 8 7
7 P8 10 5
8 P2 7 6Now, let us assume we want to test for the percent change in the number of lesions (change from preparation 1 to 2). We can accomplish this a number of ways with the t.test function.
# treat it as a one-sample problem
t.test(x = log(tb$preparation_2) - log(tb$preparation_1))
One Sample t-test
data: log(tb$preparation_2) - log(tb$preparation_1)
t = -3.1127, df = 7, p-value = 0.01702
alternative hypothesis: true mean is not equal to 0
95 percent confidence interval:
-0.4989167 -0.0681422
sample estimates:
mean of x
-0.2835295 # treat it as paired samples
(t2 = t.test(x = log(tb$preparation_2),
y = log(tb$preparation_1),
paired = TRUE))
Paired t-test
data: log(tb$preparation_2) and log(tb$preparation_1)
t = -3.1127, df = 7, p-value = 0.01702
alternative hypothesis: true mean difference is not equal to 0
95 percent confidence interval:
-0.4989167 -0.0681422
sample estimates:
mean difference
-0.2835295 These results, as interpreted using the sympercents approach, indicate a {r} round(unname(t2$estimate)*100,2) s% change from preparation 1 to preparation 2 (preparation 2 has less lesions).
Therefore, we could report the results as the following:
The second prepartion decreased lesions by approximately 28% (p = 0.017, 95% C.I [6.8%, 49.9%]).
Let’s look at the PlantGrowth data. To quote the documentation:
Results from an experiment to compare yields (as measured by dried weight of plants) obtained under a control and two different treatment conditions.
data(PlantGrowth)
pg = PlantGrowth %>%
janitor::clean_names() %>%
as_tibble()
pg# A tibble: 30 × 2
weight group
<dbl> <fct>
1 4.17 ctrl
2 5.58 ctrl
3 5.18 ctrl
4 6.11 ctrl
5 4.5 ctrl
6 4.61 ctrl
7 5.17 ctrl
8 4.53 ctrl
9 5.33 ctrl
10 5.14 ctrl
# ℹ 20 more rowsLet’s us compare the three groups using a ordinary least squares (OLS) using an Analysis of Variance (ANOVA) and making pairwise comparisons using the estimated marginal means (also called least square means).
model = lm(log(weight) ~ group,
data = pg)
flextable::as_flextable(model) %>% flextable::theme_tron()Estimate | Standard Error | t value | Pr(>|t|) | ||
|---|---|---|---|---|---|
(Intercept) | 1.610 | 0.040 | 40.583 | 0.0000 | *** |
grouptrt1 | -0.083 | 0.056 | -1.483 | 0.1496 |
|
grouptrt2 | 0.097 | 0.056 | 1.726 | 0.0958 | . |
Signif. codes: 0 <= '***' < 0.001 < '**' < 0.01 < '*' < 0.05 | |||||
Residual standard error: 0.1254 on 27 degrees of freedom | |||||
Multiple R-squared: 0.2765, Adjusted R-squared: 0.2229 | |||||
F-statistic: 5.16 on 27 and 2 DF, p-value: 0.0127 | |||||
model %>% car::Anova() %>%
broom::tidy() %>% flextable::flextable() %>% flextable::theme_tron()term | sumsq | df | statistic | p.value |
|---|---|---|---|---|
group | 0.1623884 | 2 | 5.160109 | 0.01265215 |
Residuals | 0.4248443 | 27 |
The ANOVA level effect is significant, let us make those pairwise comparisons on the sympercent scale.
library(emmeans)
emmeans(model,
pairwise ~ group,
adjust = "tukey")$emmeans
group emmean SE df lower.CL upper.CL
ctrl 1.61 0.0397 27 1.53 1.69
trt1 1.53 0.0397 27 1.45 1.61
trt2 1.71 0.0397 27 1.63 1.79
Results are given on the log (not the response) scale.
Confidence level used: 0.95
$contrasts
contrast estimate SE df t.ratio p.value
ctrl - trt1 0.0832 0.0561 27 1.483 0.3145
ctrl - trt2 -0.0968 0.0561 27 -1.726 0.2140
trt1 - trt2 -0.1800 0.0561 27 -3.209 0.0093
Results are given on the log (not the response) scale.
P value adjustment: tukey method for comparing a family of 3 estimates The output from emmeans shows the estimated marginal means and contrasts on the log scale. For the means, this is rather meaningless. However, for the contrasts, this is the sympercent. Notice, that we have a standard error for the sympercent as well.
From these results we could conclude that neither group significantly differed from control with the estimated differences being less than 10%. However, treatment 2 results in an 18% reduction in weight compared to treatment 1.
For this example, let us use the data from Plotkin et al. (2022) (data is available for download online). We will use the the outcome variable of the countermovment jump (or CMJ)
df_rct = readr::read_csv(here::here(
"blog",
"sympercentchange",
"dataset.csv"
)) %>%
janitor::clean_names() %>%
mutate(id = as.factor(code),
training_grp = case_when(
group == 0 ~ "LOAD",
group == 1 ~ "REPS"
)) %>%
dplyr::select(id, training_grp, sex, starts_with("cmj"))Rows: 38 Columns: 31
── Column specification ────────────────────────────────────────────────────────
Delimiter: ","
chr (1): SEX
dbl (30): CODE, GROUP, RF30_Pre, RF30_Post, RF50_Pre, RF50_Post, RF70_Pre, R...
ℹ Use `spec()` to retrieve the full column specification for this data.
ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.head(df_rct)# A tibble: 6 × 5
id training_grp sex cmj_pre cmj_post
<fct> <chr> <chr> <dbl> <dbl>
1 1 LOAD M 42.4 46.2
2 2 REPS F 41.4 45.7
3 3 REPS M 53.8 53.6
4 4 REPS F 38.6 43.7
5 5 LOAD F 35.3 36.6
6 6 LOAD F 31.8 37.1There are many ways the data could be analyzed. However, I’d caution that an analysis of covariance would be the most efficient option. We also have the choice of making the outcome variable the post score or the change score. Personally, I prefer the post score, it makes most sense to me1.
Let’s build the models real quick. Notice, that at the ANOVA level with type 3 sums of squares, the p-values are equivalent for the change score and post ANCOVA models.
model_post = lm(log(cmj_post)~ training_grp + log(cmj_pre) + sex,
data = df_rct)
car::Anova(model_post, type = "3") %>%
broom::tidy() %>%
flextable::as_flextable() %>% flextable::theme_tron()term | sumsq | df | statistic | p.value |
|---|---|---|---|---|
character | numeric | numeric | numeric | numeric |
(Intercept) | 0.0 | 1 | 1.4 | 0.3 |
training_grp | 0.0 | 1 | 0.0 | 0.9 |
log(cmj_pre) | 0.8 | 1 | 121.0 | 0.0 |
sex | 0.0 | 1 | 0.1 | 0.8 |
Residuals | 0.2 | 34 | ||
n: 5 | ||||
model_cng = lm(I(log(cmj_post)-log(cmj_pre))~ training_grp + log(cmj_pre) + sex,
data = df_rct)
car::Anova(model_cng, type = "3")%>%
broom::tidy() %>%
flextable::as_flextable() %>% flextable::theme_tron()term | sumsq | df | statistic | p.value |
|---|---|---|---|---|
character | numeric | numeric | numeric | numeric |
(Intercept) | 0.0 | 1 | 1.4 | 0.3 |
training_grp | 0.0 | 1 | 0.0 | 0.9 |
log(cmj_pre) | 0.0 | 1 | 1.3 | 0.3 |
sex | 0.0 | 1 | 0.1 | 0.8 |
Residuals | 0.2 | 34 | ||
n: 5 | ||||
Now, we can make a pairwise comparison using the estimated marginal means. In this case, we will average the effect, like the model, over both male and female participants
emmeans(model_post, pairwise~training_grp)$emmeans
training_grp emmean SE df lower.CL upper.CL
LOAD 3.73 0.0186 34 3.69 3.77
REPS 3.73 0.0198 34 3.69 3.77
Results are averaged over the levels of: sex
Results are given on the log (not the response) scale.
Confidence level used: 0.95
$contrasts
contrast estimate SE df t.ratio p.value
LOAD - REPS 0.00184 0.0263 34 0.070 0.9448
Results are averaged over the levels of: sex
Results are given on the log (not the response) scale. emmeans(model_cng, pairwise~training_grp)Warning in (function (object, at, cov.reduce = mean, cov.keep = get_emm_option("cov.keep"), : There are unevaluated constants in the response formula
Auto-detection of the response transformation may be incorrect$emmeans
training_grp emmean SE df lower.CL upper.CL
LOAD -0.00322 0.0186 34 -0.0410 0.0346
REPS -0.00506 0.0198 34 -0.0453 0.0352
Results are averaged over the levels of: sex
Results are given on the identity (not the response) scale.
Confidence level used: 0.95
$contrasts
contrast estimate SE df t.ratio p.value
LOAD - REPS 0.00184 0.0263 34 0.070 0.9448
Results are averaged over the levels of: sex
Note: contrasts are still on the identity scale From this we could conclude that the training modalities elicit similar CMJ responses (difference (LOAD - REPS) = 0.18%; p = 0.9448).
We could even perform an equivalence test, let us assume we want equivalence set to 10 s%.
# Confidence interval
confint(pairs(emmeans(model_post, ~ training_grp))) contrast estimate SE df lower.CL upper.CL
LOAD - REPS 0.00184 0.0263 34 -0.0517 0.0553
Results are averaged over the levels of: sex
Results are given on the log (not the response) scale.
Confidence level used: 0.95 # p-value
test(pairs(emmeans(model_post, ~ training_grp)),
delta = .1) contrast estimate SE df t.ratio p.value
LOAD - REPS 0.00184 0.0263 34 -3.728 0.0004
Results are averaged over the levels of: sex
Results are given on the log (not the response) scale.
Statistics are tests of equivalence with a threshold of 0.1
P values are left-tailed Contrary to some researcher’s beliefs, the log transformation is not magic, and we should not assume that the model fit is ideal.
As with untransformed data, I would inspect your model to ensure that the model’s assumptions are at least tenable.
One quick way to do this in R is with the performance and see packages. I’d say the plots below are not fantastic, but they aren’t horrible either!
library(see)
library(performance)
check_model(model_post)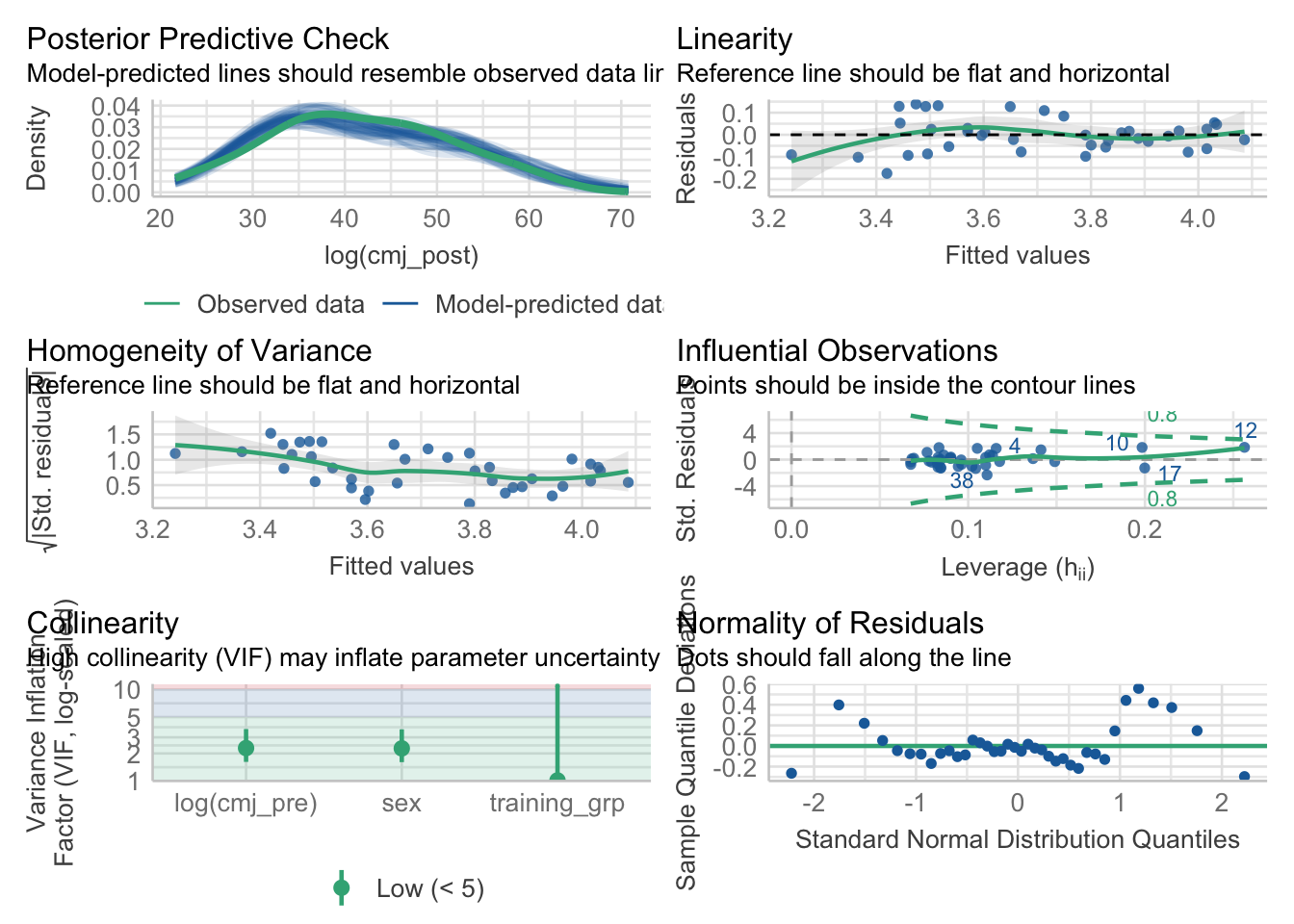
While analyzing data on its original scale is often preferred, presenting results as percentage change can be beneficial for communication and regulatory purposes2 In such cases, sympercent is a valuable tool, requiring only a natural log transformation of the raw data, and allowing regression coefficients or contrasts to be interpreted directly as sympercents, thus avoiding the asymmetry issues of traditional percentage change.
I suggest reading Frank Harrell’s blogs/books on this topic https://hbiostat.org/bbr/change#sec-change-gen↩︎
The FDA suggests that for weight loss drugs the percentage change from baseline should be reported/analyzed. I see no reason why this cannot be the sympercent.↩︎