library(palmerpenguins)
data(penguins)Sometimes as a statistician my job can be repetitive. For many projects, there may be a cluster of variables that all need to be analyzed the same way. For example, we might have a 2 x 3 factorial experiment with 12 outcome variables. A principal investigator may want to have all of them analyzed with an ANOVA, and visualized the same way in a report. Rather than using copy & paste, R can be automated (using a variety of packages) to simplify the process and make the workload (in the long run) lighter.
Data
For simplicity sake, let us use the penguins data from the palmerpenguins R package.
Formatting
Since we are doing a repetitive analysis we then want to make a “tidy” data frame with all the outcome variables. This “long” dataset can be made using the tidyverse set of packages. For this data, we will consider the experimental factors as “species” and “sex”, and consider “island” and “year” as covariates.
library(tidyverse)── Attaching core tidyverse packages ──────────────────────── tidyverse 2.0.0 ──
✔ dplyr 1.1.4 ✔ readr 2.1.4
✔ forcats 1.0.0 ✔ stringr 1.5.1
✔ ggplot2 3.4.4 ✔ tibble 3.2.1
✔ lubridate 1.9.3 ✔ tidyr 1.3.0
✔ purrr 1.0.2
── Conflicts ────────────────────────────────────────── tidyverse_conflicts() ──
✖ dplyr::filter() masks stats::filter()
✖ dplyr::lag() masks stats::lag()
ℹ Use the conflicted package (<http://conflicted.r-lib.org/>) to force all conflicts to become errorsdf_pen = penguins %>%
pivot_longer(cols = c(ends_with(c("mm","g"))), # select outcome variables
names_to = "variable", # name variables column
values_to = "y") %>% # name outcomes column
mutate(year = as.factor(year)) %>%
drop_na() # drop missing observationsNow, let us take a peek at the “long” data:
head(df_pen)# A tibble: 6 × 6
species island sex year variable y
<fct> <fct> <fct> <fct> <chr> <dbl>
1 Adelie Torgersen male 2007 bill_length_mm 39.1
2 Adelie Torgersen male 2007 bill_depth_mm 18.7
3 Adelie Torgersen male 2007 flipper_length_mm 181
4 Adelie Torgersen male 2007 body_mass_g 3750
5 Adelie Torgersen female 2007 bill_length_mm 39.5
6 Adelie Torgersen female 2007 bill_depth_mm 17.4Analysis
The analysis can then be accomplished with a nested analysis using the purrr R package and its nest function.
# Create nested data sets
df_stats1 = df_pen %>%
group_by(variable) %>%
nest() As you can see the nest function the dataset is now nested by the variable.
df_stats1# A tibble: 4 × 2
# Groups: variable [4]
variable data
<chr> <list>
1 bill_length_mm <tibble [333 × 5]>
2 bill_depth_mm <tibble [333 × 5]>
3 flipper_length_mm <tibble [333 × 5]>
4 body_mass_g <tibble [333 × 5]>Now, let us use mutate to perform our analyses by creating a number of new columns containing the output from the analyses.
For simplicity, let’s assume our request stated that they wanted 1) a ANOVA (type 3 sums of squares) and 2) a visualization of the data. I don’t necessarily recommend such a brief set of output1
library(rstatix)
Attaching package: 'rstatix'The following object is masked from 'package:stats':
filter# For our ANOVAs
df_stats2 = df_stats1 %>%
mutate(model = map(data, # build the model
~lm(y ~ species*sex+year+island,
data = .)), # dot tells map use "data" column from nested dataset
anova = map(model, # Get ANOVA table
~car::Anova(., type = "III")),
aov_summary = map(anova,
~anova_summary(.,effect.size = "pes")),
plot = map(data, # Plot raw data
~ARCmisc::gg_sum(data = ., # plot data with my bespoke package ;-)
x = species,
y = y,
group = sex,
trace = FALSE,
sum_stat = "med",
show_points = FALSE) + ggprism::theme_prism() +
scale_color_viridis_d(end = .8) +
scale_fill_viridis_d(end = .8) +
labs(y = variable, x = " ") +
theme(legend.position = "top")))Output to a Quarto Document
This part took me a long time to figure out. However, the pander R package, in my opinion, provides the best solution that works across document outputs (in my job I tend to send PDF documents to collaborators).
First, we need to import the pander package, select initial settings, and then create a function for outputting formatted tables.
library(pander)
panderOptions('big.mark', ",")
panderOptions('keep.trailing.zeros',TRUE)
table_fun = function(x, caption = "Table",digits =3){
x = as.data.frame(x) # force into data frame
if("p" %in% colnames(x)){
x = x %>%
mutate(p = scales::pvalue(p)) # format p-values
}
names(x) = pandoc.strong.return(names(x)) # output column names as bold
pander::pander(
x,
caption = caption,
digits = digits,
style = "rmarkdown",
split.tables = 300
)
}Now, we can go through a for loop to print the results of every variable in our analysis pipeline.
for (vari1 in unique(df_stats2$variable)){
# print a header
pander::pandoc.header(paste0("Analysis: ",vari1),
level = 2)
# filter out other variables
df_stats = df_stats2 %>%
filter(variable == vari1)
# ANOVA contents
pander::pandoc.p(table_fun(subset(df_stats, variable == vari1)$aov_summary[[1]],
caption = "ANOVA"))
# adding also empty lines, to be sure that this is valid Markdown
pander::pandoc.p('<br>')
# put plot on separate page if it were a PDF document
# # by adding in \newpage latex command
pander::pandoc.p('\\newpage')
# output plot
plt = subset(df_stats, variable == vari1)$plot[[1]]
print(plt)
pander::pandoc.p('\\newpage')
}Analysis: bill_length_mm
| Effect | DFn | DFd | F | p | p<.05 | pes |
|---|---|---|---|---|---|---|
| species | 2 | 323 | 261.55 | <0.001 | * | 0.618 |
| sex | 1 | 323 | 68.56 | <0.001 | * | 0.175 |
| year | 2 | 323 | 4.49 | 0.012 | * | 0.027 |
| island | 2 | 323 | 1.05 | 0.351 | 0.006 | |
| species:sex | 2 | 323 | 2.30 | 0.102 | 0.014 |

Analysis: bill_depth_mm
| Effect | DFn | DFd | F | p | p<.05 | pes |
|---|---|---|---|---|---|---|
| species | 2 | 323 | 184.523 | <0.001 | * | 0.533 |
| sex | 1 | 323 | 111.488 | <0.001 | * | 0.257 |
| year | 2 | 323 | 1.881 | 0.154 | 0.012 | |
| island | 2 | 323 | 1.184 | 0.307 | 0.007 | |
| species:sex | 2 | 323 | 0.396 | 0.674 | 0.002 |

Analysis: flipper_length_mm
| Effect | DFn | DFd | F | p | p<.05 | pes |
|---|---|---|---|---|---|---|
| species | 2 | 323 | 273.03 | <0.001 | * | 0.628 |
| sex | 1 | 323 | 28.72 | <0.001 | * | 0.082 |
| year | 2 | 323 | 26.50 | <0.001 | * | 0.141 |
| island | 2 | 323 | 4.18 | 0.016 | * | 0.025 |
| species:sex | 2 | 323 | 5.98 | 0.003 | * | 0.036 |
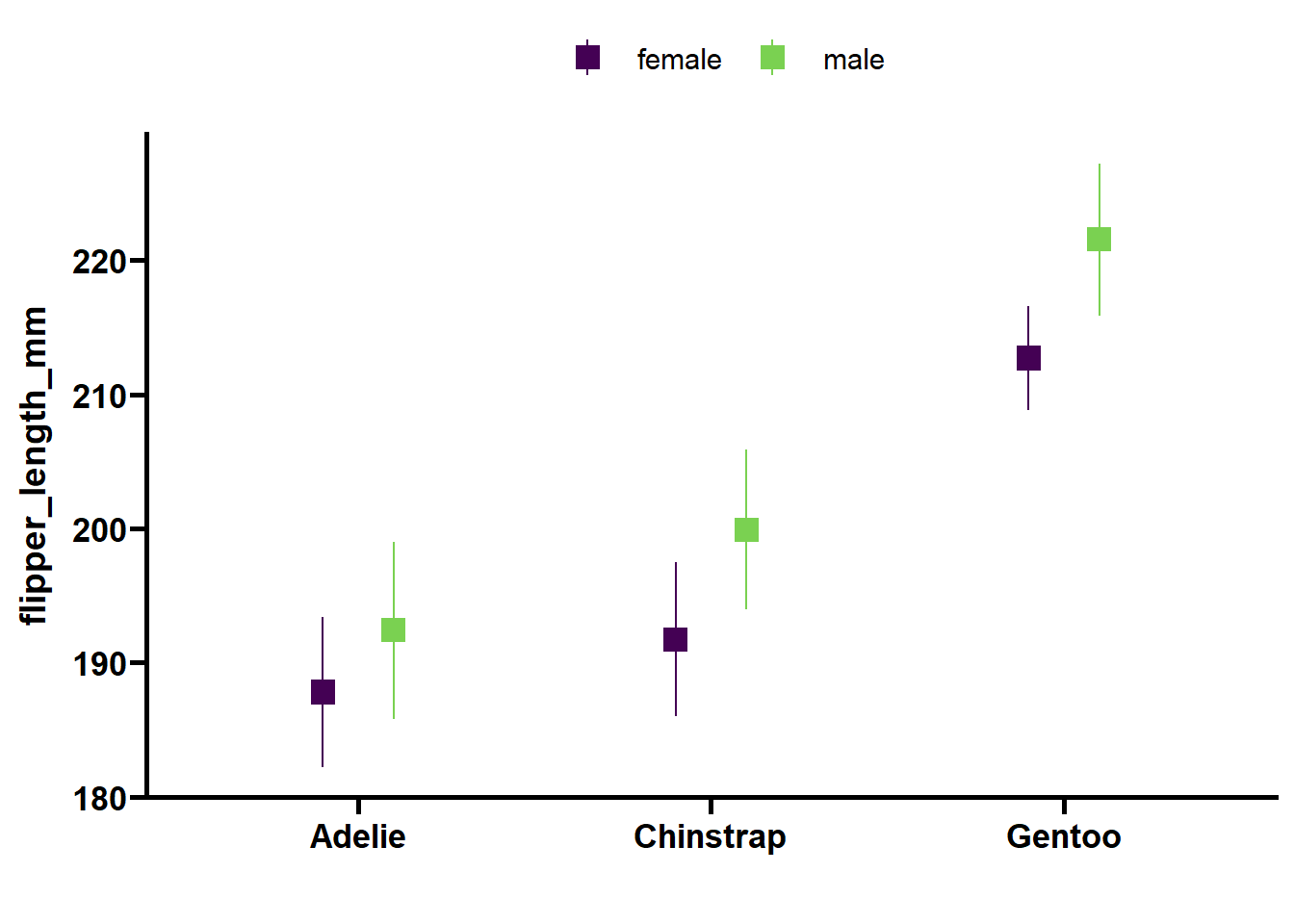
Analysis: body_mass_g
| Effect | DFn | DFd | F | p | p<.05 | pes |
|---|---|---|---|---|---|---|
| species | 2 | 323 | 188.498 | <0.001 | * | 5.39e-01 |
| sex | 1 | 323 | 171.602 | <0.001 | * | 3.47e-01 |
| year | 2 | 323 | 0.016 | 0.984 | 9.73e-05 | |
| island | 2 | 323 | 0.057 | 0.944 | 3.54e-04 | |
| species:sex | 2 | 323 | 8.655 | <0.001 | * | 5.10e-02 |
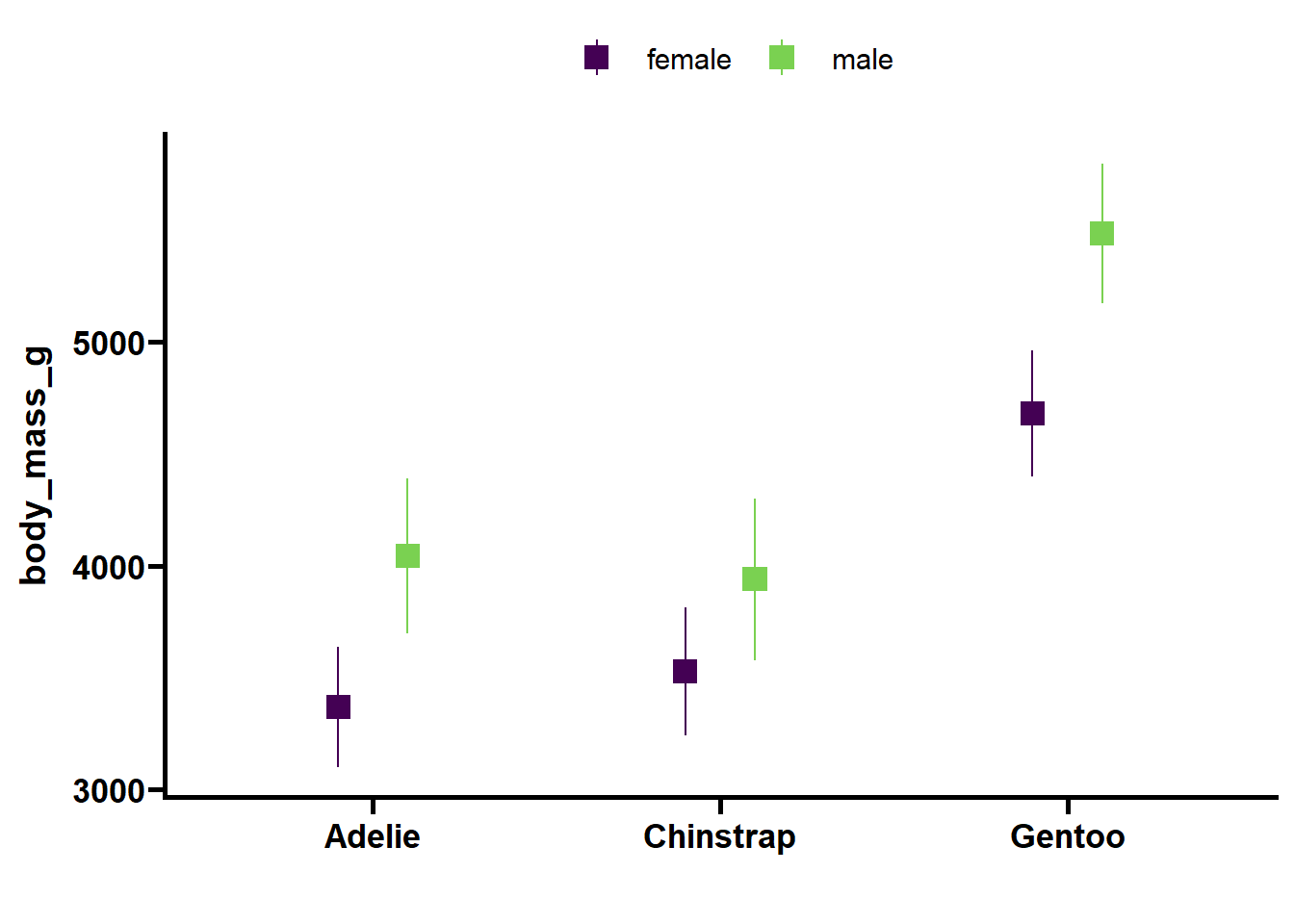
Conclusion
So, if you are ever in the situation where you have to output the same thing for multiple variables consider using some of this code as a guide. I have used this approach for over a dozen projects now, and to be quite honest I created this post so I had a publicly available version of my code. The code here may seem a bit excessive, but trust me that this will make your life easier as you start to work on the 15th version of analysis report for your collaborator where all they want you to do is change the fontsize on the y-axis for all the plots.
Last Update: 2024-01-31
Footnotes
Often the output is far more extensive and may even vary a little bit based on the variable or possibly issues with the model. Specifically, I often put the output of
check_modelfromeasystatsin the output to provide a little information on the model diagnostics.↩︎