Simulates an exact dataset (mu, sd, and r represent empirical, not population, mean and covariance matrix) from the design to calculate power
Source:R/ANOVA_exact.R
ANOVA_exact.RdSimulates an exact dataset (mu, sd, and r represent empirical, not population, mean and covariance matrix) from the design to calculate power
ANOVA_exact(
design_result,
correction = Superpower_options("correction"),
alpha_level = Superpower_options("alpha_level"),
verbose = Superpower_options("verbose"),
emm = Superpower_options("emm"),
emm_model = Superpower_options("emm_model"),
contrast_type = Superpower_options("contrast_type"),
liberal_lambda = Superpower_options("liberal_lambda"),
emm_comp
)
ANOVA_exact2(
design_result,
correction = Superpower_options("correction"),
alpha_level = Superpower_options("alpha_level"),
verbose = Superpower_options("verbose"),
emm = Superpower_options("emm"),
emm_model = Superpower_options("emm_model"),
contrast_type = Superpower_options("contrast_type"),
emm_comp,
liberal_lambda = Superpower_options("liberal_lambda")
)Arguments
- design_result
Output from the ANOVA_design function
- correction
Set a correction of violations of sphericity. This can be set to "none", "GG" Greenhouse-Geisser, and "HF" Huynh-Feldt
- alpha_level
Alpha level used to determine statistical significance
- verbose
Set to FALSE to not print results (default = TRUE)
- emm
Set to FALSE to not perform analysis of estimated marginal means
- emm_model
Set model type ("multivariate", or "univariate") for estimated marginal means
- contrast_type
Select the type of comparison for the estimated marginal means. Default is pairwise. See the emmeans package on "contrast-methods" for more details on acceptable methods.
- liberal_lambda
Logical indicator of whether to use the liberal (cohen_f^2\*(num_df+den_df)) or conservative (cohen_f^2\*den_df) calculation of the noncentrality (lambda) parameter estimate. Default is FALSE.
- emm_comp
Set the comparisons for estimated marginal means comparisons. This is a factor name (a), combination of factor names (a+b), or for simple effects a | sign is needed (a|b)
Value
Returns dataframe with simulation data (power and effect sizes!), anova results and simple effect results, plot of exact data, and alpha_level. Note: Cohen's f = sqrt(pes/1-pes) and the noncentrality parameter is = f^2*df(error)
"dataframe"A dataframe of the simulation result.
"aov_result"aovobject returned from aov_car."main_result"The power analysis results for ANOVA level effects.
"pc_results"The power analysis results for the pairwise (t-test) comparisons.
"emm_results"The power analysis results of the pairwise comparison results.
"manova_results"Default is "NULL". If a within-subjects factor is included, then the power of the multivariate (i.e. MANOVA) analyses will be provided.
"alpha_level"The alpha level, significance cut-off, used for the power analysis.
"method"Record of the function used to produce the simulation
"plot"A plot of the dataframe from the simulation; should closely match the meansplot in
ANOVA_design
Functions
ANOVA_exact2(): An extension of ANOVA_exact that uses the effect sizes calculated from very large sample size empirical simulation. This allows for small sample sizes, where ANOVA_exact cannot, while still accurately estimating power. However, model objects (emmeans and aov) are not included as output, and pairwise (t-test) results are not currently supported.
Warnings
Varying the sd or r (e.g., entering multiple values) violates assumptions of homoscedascity and sphericity respectively
Examples
## Set up a within design with 2 factors, each with 2 levels,
## with correlation between observations of 0.8,
## 40 participants (who do all conditions), and standard deviation of 2
## with a mean pattern of 1, 0, 1, 0, conditions labeled 'condition' and
## 'voice', with names for levels of "cheerful", "sad", amd "human", "robot"
design_result <- ANOVA_design(design = "2w*2w", n = 40, mu = c(1, 0, 1, 0),
sd = 2, r = 0.8, labelnames = c("condition", "cheerful",
"sad", "voice", "human", "robot"))
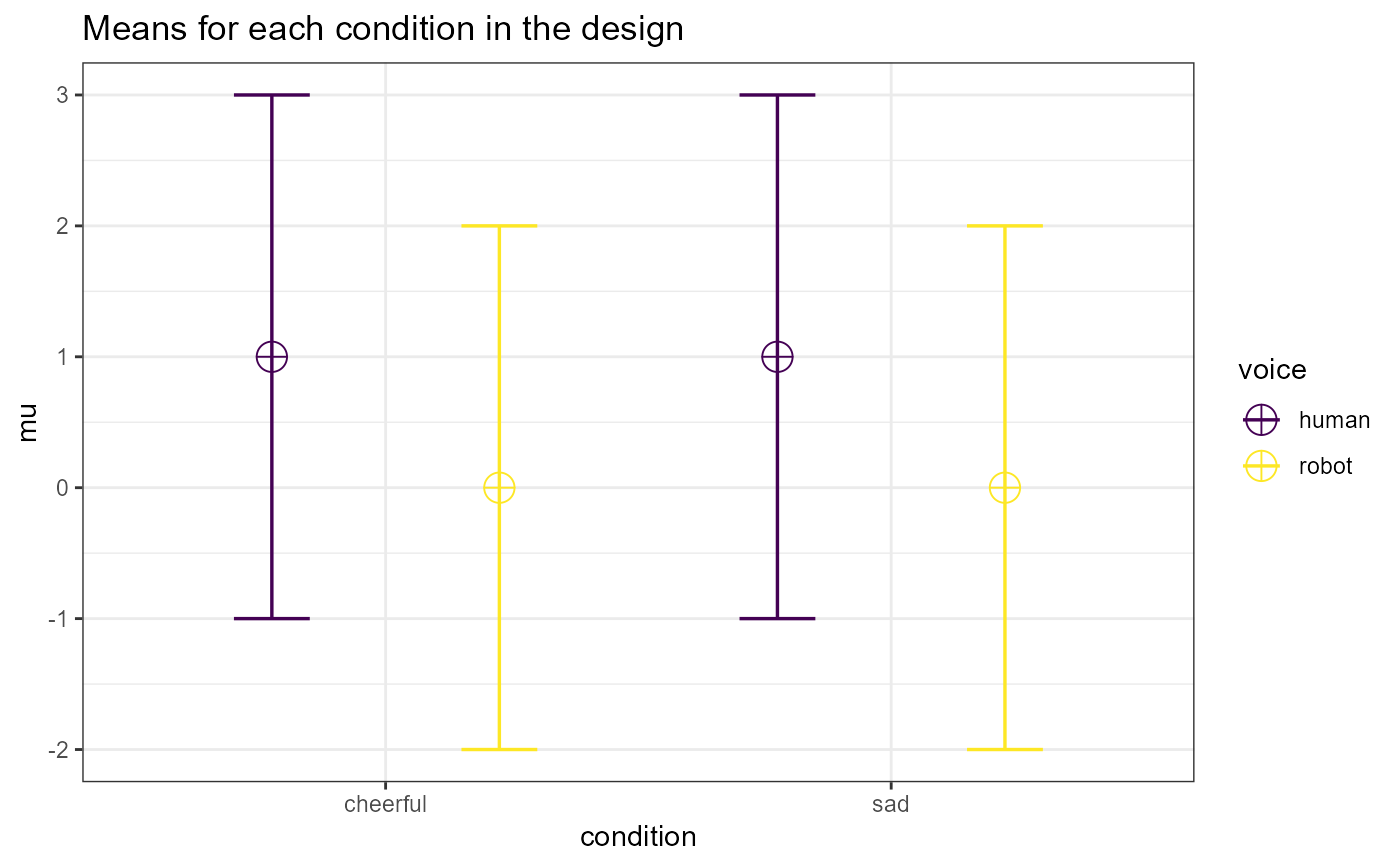 exact_result <- ANOVA_exact(design_result, alpha_level = 0.05)
#> Power and Effect sizes for ANOVA tests
#> power partial_eta_squared cohen_f non_centrality
#> condition 5 0.0000 0.0000 0
#> voice 100 0.5618 1.1323 50
#> condition:voice 5 0.0000 0.0000 0
#>
#> Power and Effect sizes for pairwise comparisons (t-tests)
#> power
#> p_condition_cheerful_voice_human_condition_cheerful_voice_robot 99.82
#> p_condition_cheerful_voice_human_condition_sad_voice_human 5.00
#> p_condition_cheerful_voice_human_condition_sad_voice_robot 99.82
#> p_condition_cheerful_voice_robot_condition_sad_voice_human 99.82
#> p_condition_cheerful_voice_robot_condition_sad_voice_robot 5.00
#> p_condition_sad_voice_human_condition_sad_voice_robot 99.82
#> effect_size
#> p_condition_cheerful_voice_human_condition_cheerful_voice_robot -0.79
#> p_condition_cheerful_voice_human_condition_sad_voice_human 0.00
#> p_condition_cheerful_voice_human_condition_sad_voice_robot -0.79
#> p_condition_cheerful_voice_robot_condition_sad_voice_human 0.79
#> p_condition_cheerful_voice_robot_condition_sad_voice_robot 0.00
#> p_condition_sad_voice_human_condition_sad_voice_robot -0.79
exact_result <- ANOVA_exact(design_result, alpha_level = 0.05)
#> Power and Effect sizes for ANOVA tests
#> power partial_eta_squared cohen_f non_centrality
#> condition 5 0.0000 0.0000 0
#> voice 100 0.5618 1.1323 50
#> condition:voice 5 0.0000 0.0000 0
#>
#> Power and Effect sizes for pairwise comparisons (t-tests)
#> power
#> p_condition_cheerful_voice_human_condition_cheerful_voice_robot 99.82
#> p_condition_cheerful_voice_human_condition_sad_voice_human 5.00
#> p_condition_cheerful_voice_human_condition_sad_voice_robot 99.82
#> p_condition_cheerful_voice_robot_condition_sad_voice_human 99.82
#> p_condition_cheerful_voice_robot_condition_sad_voice_robot 5.00
#> p_condition_sad_voice_human_condition_sad_voice_robot 99.82
#> effect_size
#> p_condition_cheerful_voice_human_condition_cheerful_voice_robot -0.79
#> p_condition_cheerful_voice_human_condition_sad_voice_human 0.00
#> p_condition_cheerful_voice_human_condition_sad_voice_robot -0.79
#> p_condition_cheerful_voice_robot_condition_sad_voice_human 0.79
#> p_condition_cheerful_voice_robot_condition_sad_voice_robot 0.00
#> p_condition_sad_voice_human_condition_sad_voice_robot -0.79